The three-dimensional (3D) genome structure and epigenetic state play critical roles in transcriptional regulation. Comprehensive analyses of these features facilitate understanding the molecular basis underlying cell fate determination and disease mechanisms, ultimately advancing precision medicine. Currently, most 3D genome mapping approaches rely on bulk-cell data, capturing averaged cellular states and lacking single-cell resolution. On the other hand, although single-cell multi-omics methods that combine scRNA-seq and scATAC-seq simultaneously profile transcriptional activity and chromatin accessibility, they do not provide chromatin interaction data. Similarly, methods combining scRNA-seq with scHi-C, while capable of capturing chromatin interactions and gene expression, fail to accurately resolve specific cis-regulatory element-promoter interactions. Therefore, developing technologies capable of simultaneously capturing transcriptional, epigenetic, and 3D structural information at single-cell resolution is essential for elucidating the regulatory mechanisms of gene expression.
To fill this technological gap, Dr. Ruan's group previously developed ChIATAC [1], a method capable of simultaneously detecting chromatin interactions and chromatin accessibility at the bulk-cell level. Building upon this, the group further developed the single-cell multi-omic technology ChAIR [2].
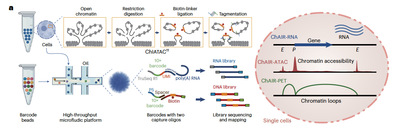
Utilizing droplet-based microfluidics, ChAIR simultaneously captures chromatin accessibility (ChAIR-ATAC), 3D chromatin structure (ChAIR-PET), and gene expression information (ChAIR-RNA) in individual cells, producing datasets of over 100,000 single cells per experiment. Specifically, ChAIR-RNA identifies cell types, ChAIR-ATAC detects chromatin accessibility, and ChAIR-PET captures long-range chromatin interactions. Together, these modules dissect cell-type-specific 3D epigenomic regulatory networks and their relationship with transcriptional activation. ChAIR significantly surpasses existing 3D genomic methods in throughput and resolution, allowing precise identification of transcription-associated chromatin loops and higher-order structures at a cell-type-specific level, thus advancing mechanistic studies on the role of the 3D epigenome in gene regulation.
Leveraging ChAIR’s high resolution, Dr. Ruan’s group analyzed tri-omic single-cell data from 35,515 mouse Patski cells and 222,698 mouse brain cells. They demonstrated that chromatin interactions between target genes and distal cis-regulatory elements are established prior to transcriptional activation during mitosis and cell differentiation. Such pre-established interactions are followed by increased chromatin accessibility and subsequent transcriptional activation of target genes, indicating a potential causal 3D epigenomic mechanism in regulating gene expression.
Furthermore, by integrating ChAIR-RNA data with corresponding spatial transcriptomic data, the study revealed spatially specific 3D epigenomic and transcriptomic features across different tissues. These findings were published in Nature Methods on April 29, 2025, in the article titled Tri-omic single-cell mapping of the 3D epigenome and transcriptome in whole mouse brains throughout the lifespan.
[1] Chai, H. et al. ChIATAC is an efficient strategy for multi-omics mapping of 3D epigenomes from low-cell inputs. Nat. Commun. 14, 213 (2023).
[2] Chai, H., Huang, X., Xiong, G. et al. Tri-omic single-cell mapping of the 3D epigenome and transcriptome in whole mouse brains throughout the lifespan. Nat Methods (2025). https://doi.org/10.1038/s41592-025-02658-7
Dr. Haoxi Chai, Research Assistant Professor and Dr. Xingyu Huang, postdoctoral researcher from Dr. Ruan’s group, are co-first authors. Dr. Yijun Ruan is the corresponding author. The research was supported by the National Natural Science Foundation of China.
Link to original article: https://www.nature.com/articles/s41592-025-02658-7