On August 26, 2025, the research group led by Professor Shixian Lin from Life Sciences Institute published a research article in Nature Communications, titled Nascent liver proteome reveals enzymes and transcription regulators under physiological and alcohol exposure conditions.
The liver performs hundreds of critical biological functions, supported by the highly dynamic regulation of its proteome. Under alcohol exposure, the dysregulation of liver protein homeostasis is a core cause of alcoholic liver disease (e.g., fatty liver, hepatitis, cirrhosis). Traditional proteomic techniques typically provide only a static panoramic view of proteins, making it difficult to capture the crucial nascent proteins (Nascent Proteome) involved in the onset and progression of disease. Therefore, there is an urgent need for a new technology that can accurately target the liver in living animals and capture nascent protein information in real-time to gain a deeper understanding of the molecular mechanisms of liver disease.
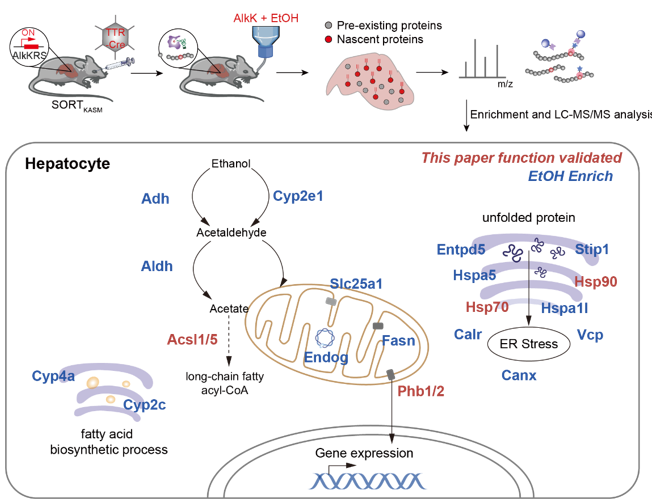
Figure 1. Labeling, identification, and functional analysis of the nascent proteome in the liver under alcohol exposure conditions.
To address this challenge, the study established a universal strategy for liver-specific labeling and identification of the nascent proteome in living animals, named SORT-AC. This strategy ingeniously combines genetic code expansion technology, genetically edited mouse models, and a viral delivery system: 1) First, an aminoacyl-tRNA synthetase (AlkKRS) capable of efficiently recognizing the non-canonical amino acid (AlkK). 2) To preferentially enrich membrane proteins, the tRNA's anticodon was engineered to recognize several natural amino acids (K, A, S, M) that are abundant in transmembrane proteins. 3) A transgenic mouse line with the conditional expression labeling system, SORT-KASM, was constructed. Finally, the gene for the hepatocyte-specific TTR-Cre recombinase was delivered into the mice via a targeted adeno-associated virus (AAV), and AlkK was added to the mice's drinking water, successfully achieving the labeling and identification of the hepatocyte nascent proteome under physiological or pathological conditions.
The study successfully identified a number of proteins involved in amino acid, lipid, and small-molecule metabolism. These proteins were particularly enriched in the cell membrane and key organelles like the mitochondria and endoplasmic reticulum, revealing the molecular basis for how the liver maintains homeostasis. Notably, the research revealed that Phb1/2 act as mitochondrial membrane-associated transcriptional co-regulators that play a crucial role in the transcriptional regulation of alcohol metabolism-related genes. Additionally, the study found that the fatty acid-activating enzymes Acsl1/5 are key enzymes in ethanol-induced lipid accumulation in the liver. Intervention with their inhibitors significantly reduced ethanol-induced lipid accumulation in both cells and mouse livers.
The first authors of the article include Jiayu Gu and Lihui Lao. The corresponding authors are Professor Shixian Lin, Yulin Chen, and Ying Yuan.
Article link: https://www.nature.com/articles/s41467-025-63212-9
Lab website: /25713/list.htm