On August 22, 2025, Dr. Yijun Ruan and Dr. Haoxi Chai published a commissioned review article in Trends in Genetics titled “3D genome sequencing technology in its mid-teens: past, present, and future.” In this article, they comprehensively summarized the development of DNA high-throughput sequencing technologies and analytical tools for deciphering 3D genome architecture, outlined recent advances in the field, and provided perspectives on future directions.

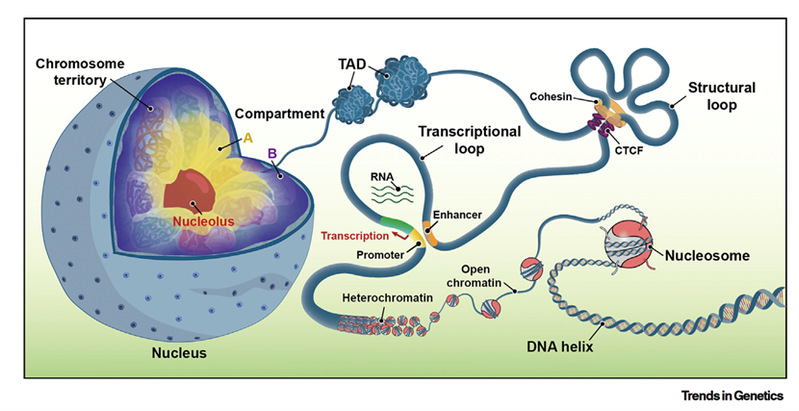
The genome is packaged with nuclear proteins and RNAs into a complex struc-ture known as chromatin. Its dynamic organization influences genome functions and nuclear properties. Since 2009, high-throughput DNA sequencing methods such as Hi-C and ChIA-PET have pioneered genome-wide mapping of chromatin folding architectures and have given rise to the field of three-dimensional (3D) genome biology. Now, after 15 years of development, this field has experienced a remarkable growth and is still expanding rapidly. It is significantly deepening our understanding of how genome organization affects nuclear functions in various biological systems. The article primarily introduces two categories of 3D genome sequencing strategies: enrichment-based and non-enrichment-based. Enrichment-based approaches are mainly applied to investigate chromatin interactions mediated by proteins, among open chromatin regions, or at specific genomic loci, whereas non-enrichment approaches are designed to provide an unbiased view of the global 3D genome architecture. In addition, the review systematically discusses single-cell multi-omics technologies for 3D genome analysis and highlights the new perspectives they may offer in studies of disease, development, and evolution.
This work was supported by the National Natural Science Foundation of China.
Link: https://doi.org/10.1016/j.tig.2025.07.010